Omics Discovery Index is an integrated and open source platform facilitating the access and dissemination of omics datasets. It provides a unique infrastructure to integrate datasets coming from multiple omics studies, including at present proteomics, genomics, transcriptomics and metabolomics.
OmicsDI stores metadata coming from the public datasets from every resource using an efficient indexing system, which is able to integrate different biological entities including genes, proteins and metabolites with the relevant life science literature. OmicsDI is updated daily, as new datasets get publicly available in the contributing repositories.
Omics Data Submission
The increasing role of huge datasets in scientific research has important implications for the way the research is conducted, for the way it should be organized and funded, and for the training of new researchers. However, the advances in biomedical research depend on scientists’ ability to consult and use all available data, independently from where they were originally produced: data sharing on a global scale is the best way to ‘advance science for the public good’.
The assumption underlying this policy is that the more scientists are allowed to access the same sets of data, the more those data will be used to produce new knowledge about biological phenomena.
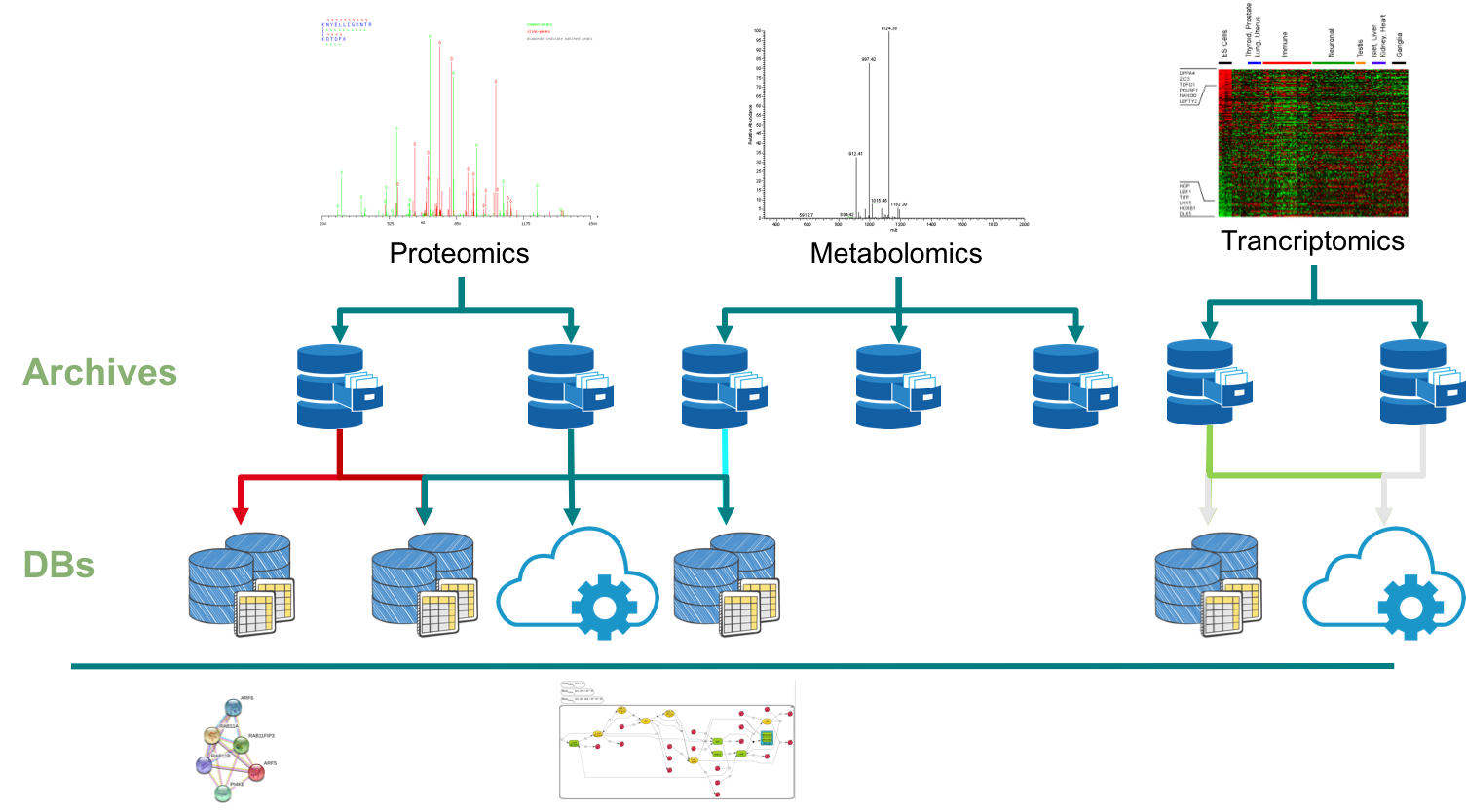
Figure 1: Current Schema of BioMedical Data Distribution/Dissemination
Figure 1 shows how data is produced/stored and distributed in biomedical research. The result of an omics experiment (e.g Proteomics or Metabolomics)
is submitted to a public Archive (e.g PRIDE or Metaboligths). These Data Archives provide a common interface for submission,
validation and downloading of the original results/data. Importantly, each individual repository/archive define three major
components to guide the submission process:
- The metadata guidelines including Standards and Ontologies to define/describe a dataset and corresponding components (e.g. samples, instruments).
- File formats to store and handle the underlying data in the Dataset.
- The submission guidelines define how to submit and retrieve the data from the repository.
In recent years the number of databases and archives has growth in all omics fields [1]. For example,
in Proteomics the results of a Mass spectrometry experiment can be submitted to four different databases members of
ProteomeXchange: PeptideAtlas/PASSEL, PRIDE, MassIVE, jPOST. In addition, each omics field
has developed and grown independently of the other fields including their metadata specifications, file formats, and submission
guidelines. For this reason, most of the Data Archives are field specific (e.g. Metabolomics - Metaboligths, Metabolomics Workbench).
Omics Data Dissemination
After the data is submitted to a formal Archive, Knowledge Base Databases
(BDs) reuse part of the public data to respond to specific questions (e.g. Gene Expression Profiles - ExpressionAtlas). The
number of these DBs has growth in recent years. For example, Table 1 shows the list of Protein Expression Databases [1] that include
peptide sequences, post-translational modifications, expression profiles.
Table 1: Proteomics Knowledge Base Databases (BDs) including information about Protein Expression informations (e.g
Peptides Sequences, Post-Translational Modifications).
| Resource | URL | Publication |
|---|---|---|
| Cancer Mutant Proteome Database | http://cgbc.cgu.edu.tw/cmpd/ | Nucleic Acids Res. 2015. |
| MOPED | https://www.proteinspire.org/MOPED/ | Nucleic Acids Res. 2015. |
| ProteomicsDB | https://www.proteomicsdb.org/ | Nature. 2014 |
| MaxQB | http://maxqb.biochem.mpg.de/mxdb/ | Mol Cell Proteomics. 2012 |
| GPMDB | http://gpmdb.thegpm.org/ | |
| COPaKB | http://www.heartproteome.org/copa/Default.aspx | Circ Res. 2013 |
| paxDB | http://pax-db.org/#!home | Mol Cell Proteomics. 2012 |
| Human Proteinpedia | http://www.humanproteinpedia.org/ | Curr Protoc Bioinformatics. 2013 |
| Human Proteome Map | http://www.humanproteomemap.org/ | Nature. 2014 |
OmicsDI Vision

All these databases/repositories have created a complex and distributed scenario where the data can
be submitted into different Archives and reused in multiple and different DBs. The development of tools
facilitating data sharing and able to handle this complexity is a great challenge in itself.
In this context, we introduce here the Omics Discovery Index, an open-source platform facilitating the access, discovery, and dissemination of omics datasets. OmicsDI provides a unique infrastructure to integrate datasets coming from multiple omics fields, including at present proteomics, genomics, metabolomics, and transcriptomics.
To date, eleven resources have agreed on a common metadata structure framework and exchange format, and have contributed to OmicsDI, including:
Proteomics: The PRoteomics IDEntifications (PRIDE) database, PeptideAtlas, the Mass spectrometry Interactive Virtual Environment (MassIVE) and the Global Proteome Machine Database (GPMDB).Metabolomics: MetaboLights, the Global Natural Products Social Molecular Networking project (GNPS), MetabolomeExpress, and the Metabolomics Workbench.The major European Genome-Phenome Archive (EGA).Transcriptomics: ArrayExpress and Expression Atlas.
OmicsDI stores biological and technical metadata coming from the public datasets available in every resource,
using an efficient indexing system, which is able to integrate differently
biological entities including genes, transcripts, proteins and metabolites with the relevant scientific literature.
References
- [1] Perez‐Riverol, Yasset, et al. “Making proteomics data accessible and reusable: current state of proteomics databases and repositories.” Proteomics 15.5-6 (2015): 930-950.