The main goal of the Omics Discovery Index is to provide a platform for searching and linking omics public data.
OmicsDI has implemented a unique and novel Search Engine for omics datasets including public and protected data.
The OmicsDI Search Box

Figure 1: OmicsDI Search Box
The OmicsDI Search Box is the main component to searching in OmicsDI. The user can type a set of keywords that will enable the system
to find the datasets containing those keywords.
If the user uses double quote "breast cancer" in their search the system will try to find the exact sentence in the datasets.The OmicsDI Search Box provides a unique auto-complete feature that enables user to select sentence after typing a subset of keywords. For example,
Figure 2 shows all sentences/phrases in OmicsDI containing the words breast cancer.
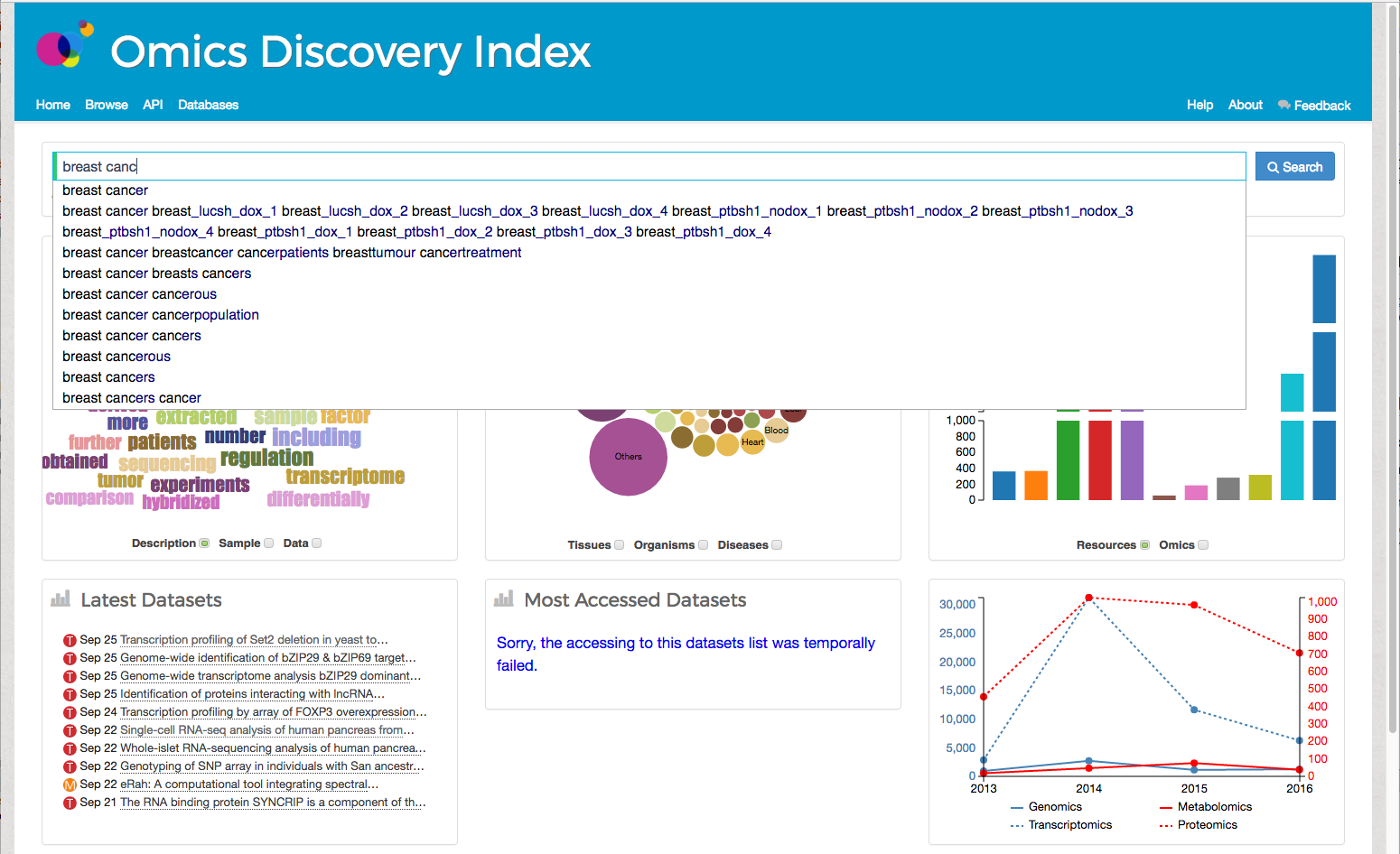
Figure 2: OmicsDI Search Box with Auto-complete
Query Syntax
When the user types any text in OmicsDI Search Box, the input is translated into an Apache Lucene query that is then executed
to get the search results. The actual query executed is generated following the typical Apache Lucene query syntax in order to
provide a generic approach avoiding complex query rearrangements.
Multiple search terms separated by white spaces are combined by default in AND logic. Therefore an input text containing for example
glutathione transferase is treated as glutathione AND transferase and only entries having both terms will be found. The default order
of results is based on their relevance, i.e. the proximity of the terms in the entries.
Table 1: Overview of some useful query syntax elements is presented.
| Element | Meaning | Usage | Example | Notes |
|---|---|---|---|---|
AND |
In addition to | term1 AND term2 | glutathione AND transferase | Matches entries where both glutathione and transferase occur. |
OR |
Equivalence | term1 OR term2 | glutathione OR transferase | Matches entries where either glutathione or transferase occur. |
NOT |
Exclusion | term1 NOT term2 | coding NOT fragment | Matches entries containing coding but not fragment. |
* |
Wildcard | partialTerm* | gluta* | Matches for instance glutathione, glutamate, glutamic. |
| ” “ | Exact match | “quoted text” | “x-ray diffraction” | Exact matching for entries containing x-ray diffraction. |
( ) |
Grouping | (text) | (reductase OR transferase) AND glutathione | |
Field: |
Field-specific search | fieldId:term | description:dopamine | Matches for a field description containing dopamine. |
Escaping special characters
The following characters within queries require to be escaped (using a ‘ \ ‘ before the character to escape) in order to be correctly interpreted:
+ - & | ! ( ) { } [ ] ^ " ~ * ? : \ /Since Apache Lucene supports regular expression searches (matching a pattern between forwarding slashes) the forward slash ‘ / ’ has become a special character to be escaped. For example to search for
cancer/testis use the query cancer\/testis. If special characters are not escaped the actual query performed may be different from what expected.
Query examples
Following the aforementioned query syntax, users can easily search and filter results according to data content and characteristics. A few examples of queries that can be performed using EBI Search are listed below.
- Search for insulin receptor.
- Search Datasets that identified P07900
Searching using Biological Evidence
The OmicsDI Search Box allows the end-users to search data using biological evidence such as the list of the proteins identified in the proteomics experiment or the metabolites
reported in the Metabolomics experiment. For example (Figure 3), if the user searches for 3-methyl-2-oxobutanoic in the resource it will find one dataset in Metaboligths and five in Metabolome workbench
that identified the current molecule.
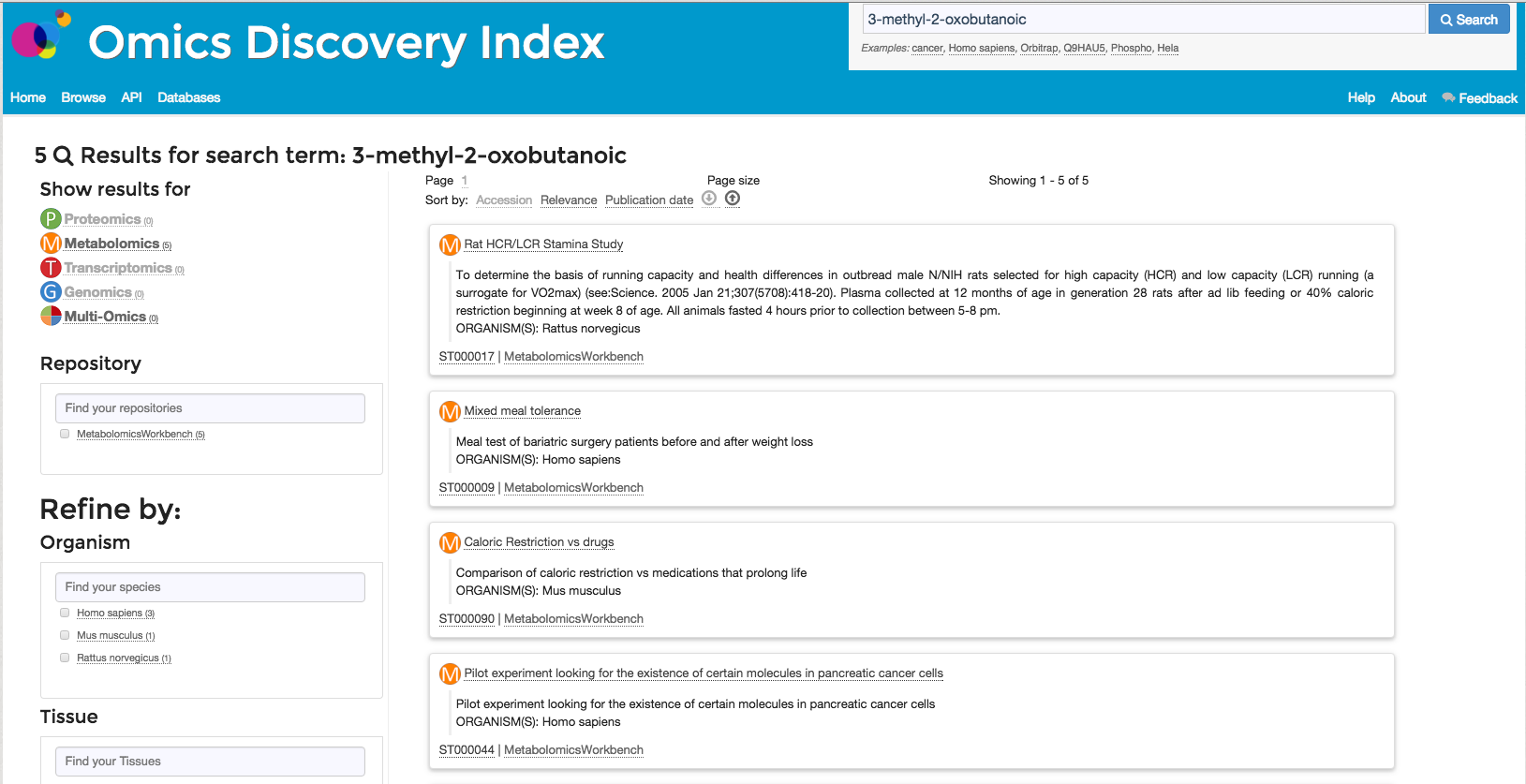
Figure 3: Search for Biological evidences 3-methyl-2-oxobutanoic
The final search results are shown in the browser page including Refine Filters. Read More Here.